Clinically applicable deep learning for diagnosis and referral in retinal disease
13 Aug 2018
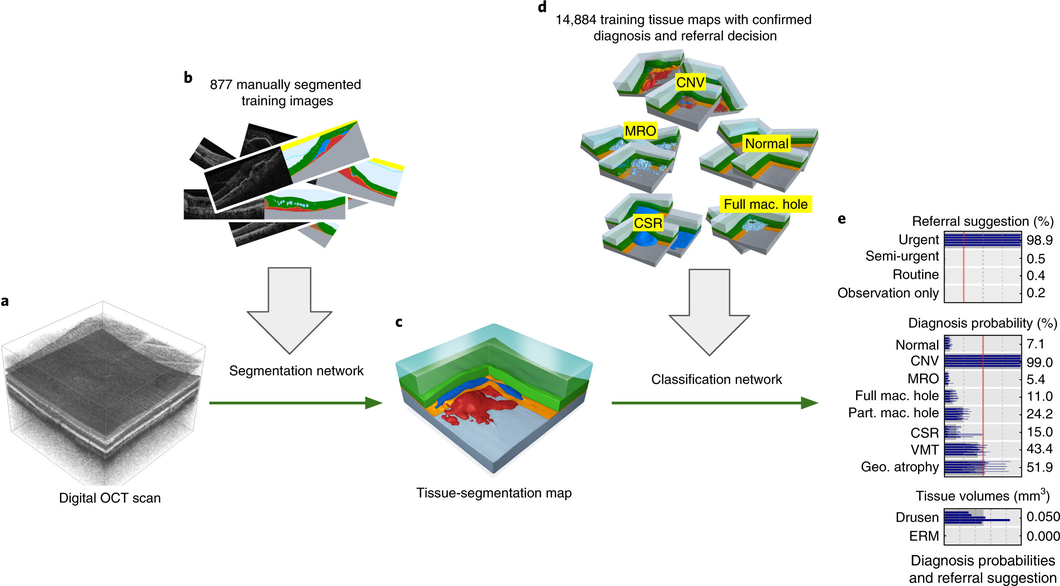
Our proposed two-stage architecture trained with sparse segmentations and diagnosis and referral labels.
We propose a two-stage architecture that consists of first mapping the original (noisy) 3D Optical Coherence Tomography (OCT) scan to multiple tissue-segmentation hypotheses, and consequently using a classification network on these tissue maps to infer diagnosis and referral probabilities. On these tasks we achieve expert-level results or better. One benefit of the two-stage architecture is that it allows for much quicker transfer to different device types, as demonstrated in the paper.
Nature Medicine article
Open-access link
BBC article

Results on the patient referral decision. Performance on an independent test set of n=997 patients (252 urgent, 230 semi-urgent, 266 routine, 249 observation only). a, ROC diagram for urgent referral (for choroidal neovascularization (CNV)) versus all other referrals. The blue ROC curve is created by sweeping a threshold over the predicted probability of a particular clinical diagnosis. Points outside the light blue area correspond to a significantly different performance (95% confidence level, using a two-sided exact binomial test). The asterisk denotes the performance of our model in the ‘balanced performance’ setting. Filled markers denote experts’ performance using OCT only; empty markers denote their performance using OCT, fundus image and summary notes. Dashed lines connect the two performance points of each expert. b, Confusion matrices with patient numbers for referral decision for our framework and the two best retina specialists. These show the number of patients for each combination of gold standard decision and predicted decision. The numbers of correct decisions are found on the diagonal. Wrong decisions due to overdiagnosis are in the bottom-left triangle, and wrong decisions due to underdiagnosis are in the top-right triangle. c, Total error rate (1 − accuracy) on referral decision. Values outside the light-blue area (3.9–7.3%) are significantly different (95% confidence interval, using a two-sided exact binomial test) to the framework performance (5.5%). AUC, area under curve.
Jeffrey De Fauw, Joseph R. Ledsam, Bernardino Romera-Paredes, Stanislav Nikolov, Nenad Tomasev, Sam Blackwell, Harry Askham, Xavier Glorot, Brendan O’Donoghue, Daniel Visentin, George van den Driessche, Balaji Lakshminarayanan, Clemens Meyer, Faith Mackinder, Simon Bouton, Kareem Ayoub, Reena Chopra, Dominic King, Alan Karthikesalingam, Cían O. Hughes, Rosalind Raine, Julian Hughes, Dawn A. Sim, Catherine Egan, Adnan Tufail, Hugh Montgomery, Demis Hassabis, Geraint Rees, Trevor Back, Peng T. Khaw, Mustafa Suleyman, Julien Cornebise, Pearse A. Keane & Olaf Ronneberger
Abstract
The volume and complexity of diagnostic imaging is increasing at a pace faster than the availability of human expertise to interpret it. Artificial intelligence has shown great promise in classifying two-dimensional photographs of some common diseases and typically relies on databases of millions of annotated images. Until now, the challenge of reaching the performance of expert clinicians in a real-world clinical pathway with three-dimensional diagnostic scans has remained unsolved. Here, we apply a novel deep learning architecture to a clinically heterogeneous set of three-dimensional optical coherence tomography scans from patients referred to a major eye hospital. We demonstrate performance in making a referral recommendation that reaches or exceeds that of experts on a range of sight-threatening retinal diseases after training on only 14,884 scans. Moreover, we demonstrate that the tissue segmentations produced by our architecture act as a device-independent representation; referral accuracy is maintained when using tissue segmentations from a different type of device. Our work removes previous barriers to wider clinical use without prohibitive training data requirements across multiple pathologies in a real-world setting.